ex10_section
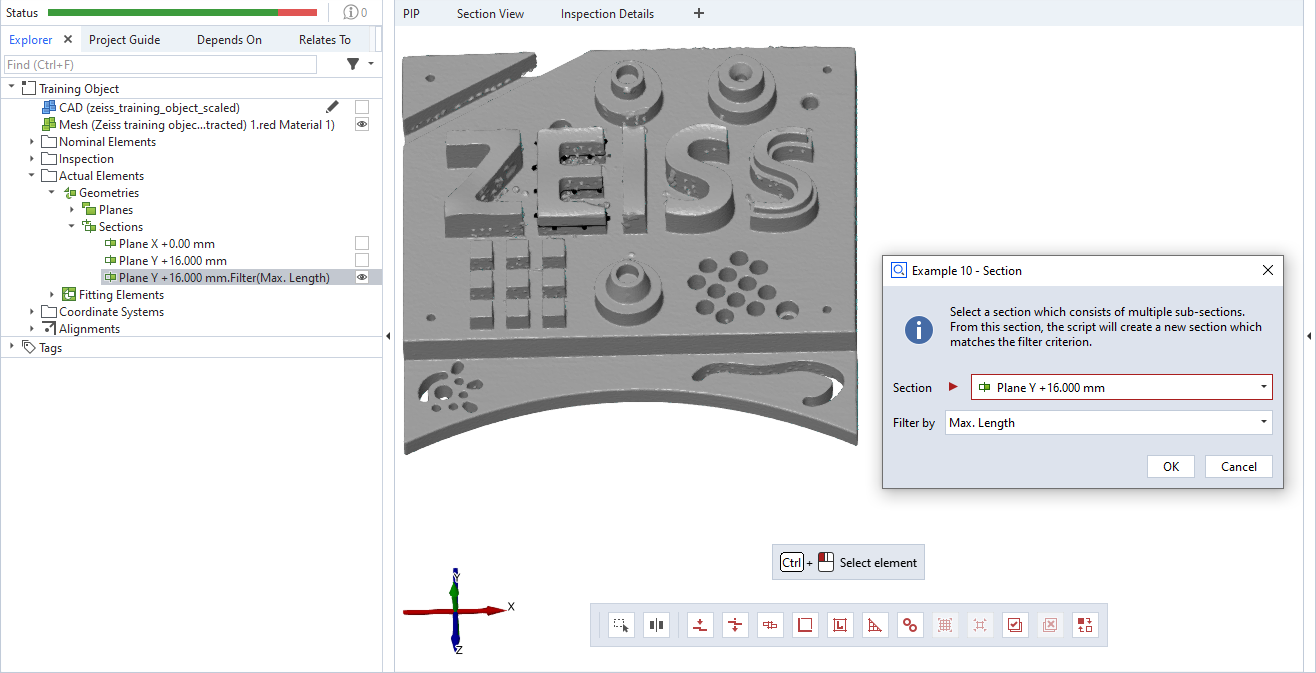
This is an example for a scripted ‘section’ element. The dialog allows to select an existing section, which may consist of multiple sub sections. The script creates a new section from the existing one, with the selected filter criterion.
Note
Please see offset_point_v2.md for a complete scripted elements example with detailed description.
Functions for calculating the section
1def calc_section_length(verts, indices=None):
2 '''Calculate length of (sub-)section'''
3 if indices is None:
4 indices = range(len(verts))
5 l = 0
6
7 # Length: Sum of Euclidean distance between each two adjacent vertices of the curve
8 for i, j in zip(indices, indices[1:]):
9 l += la.norm(verts[j] - verts[i])
10 return l
11
12
13def get_sub_sections(base_curve, stage=0):
14 """Separate section curve into sub-sections by checking if a point is connected to another point
15
16 Returns:
17 array of curves
18 """
19 curve_coords = np.array(base_curve.data.coordinate)[stage]
20 curve_normals = np.array(base_curve.data.normal)[stage]
21 sub_sections = []
22 start_index = 0
23 for end_index, conn in enumerate(base_curve.scanline_point_connection):
24 if conn != "connected":
25 sub_sections.append((curve_coords[start_index:end_index + 1], curve_normals[start_index:end_index + 1]))
26 start_index = end_index + 1
27 return sub_sections
28
29
30def filter_by_length(sub_sections, mode):
31 '''Filter an array of curves by length and return the curve matching the filter criterion'''
32 max_len = 0
33 min_len = 0
34 r_min = None
35 r_max = None
36
37 # For all sub-sections, calculate min. and max. length
38 # and save the vertices and normals of both the shortest/longest curve
39 for verts, normals in sub_sections:
40 ssl = calc_section_length(verts)
41 if max_len < ssl:
42 max_len = ssl
43 r_max = verts, normals
44 if min_len == 0 or min_len > ssl:
45 min_len = ssl
46 r_min = verts, normals
47 if mode.lower() == "max. length":
48 return r_max
49 else:
50 return r_min
Dialog and calculation functions
1def dialog(context, params):
2 #[...]
3
4def calculation(context, params):
5 '''Scripted section calculation function'''
6 valid_results = False
7
8 # Check if filter mode is defined
9 if params["i_mode"].lower() in ["min. length", "max. length"]:
10 f = filter_by_length
11 else:
12 raise ValueError(f"Unknown mode: {params['i_mode']}")
13
14 for stage in context.stages:
15 # Get selected section from stage
16 section = params["i_elem"].in_stage[stage]
17
18 # Get sub-sections
19 sub_sections = get_sub_sections(section, stage)
20
21 # Apply filter
22 sub_section = f(sub_sections, params["i_mode"])
23
24 # Get vertices and normals of resulting sub-section
25 verts, normals = sub_section
26
27 data = [{"points": [gom.Vec3d(*v) for v in verts], "normals": [gom.Vec3d(*n) for n in normals]}]
28
29 try:
30 context.result[stage] = {"curves": data}
31 context.data[stage] = {"ude_mykey": "Example 10"}
32 except Exception as error:
33 context.error[stage] = str(error)
34 else:
35 valid_results = True
36
37 return valid_results